Overview
High throughput quantitative analysis of microscopy images presents a challenge due to the complexity of the image content and the difficulty to retrieve precisely annotated datasets. In this repository we introduce a weakly-supervised MICRoscopy Analysis neural network (MICRA-Net) that can be trained on a simple main classification task using image-level annotations to solve multiple more complex auxiliary tasks, such as segmentation, detection, and enumeration.
MICRA-Net relies on the latent information embedded within a trained model to achieve performances similar to state-of-the-art fully-supervised learning. This learnt information is extracted from the network using gradient class activation maps, which are combined to generate precise feature maps of the biological structures of interest.
The source code is publicly available on GitHub.
Paper
The paper is available here.
Trained models
This section contains the link to the models that were trained for each of the presented datasets of the paper. The user may refer to the figures from the paper. The user may click on the images to download the zoo models associated with each datasets or click on the dataset name.
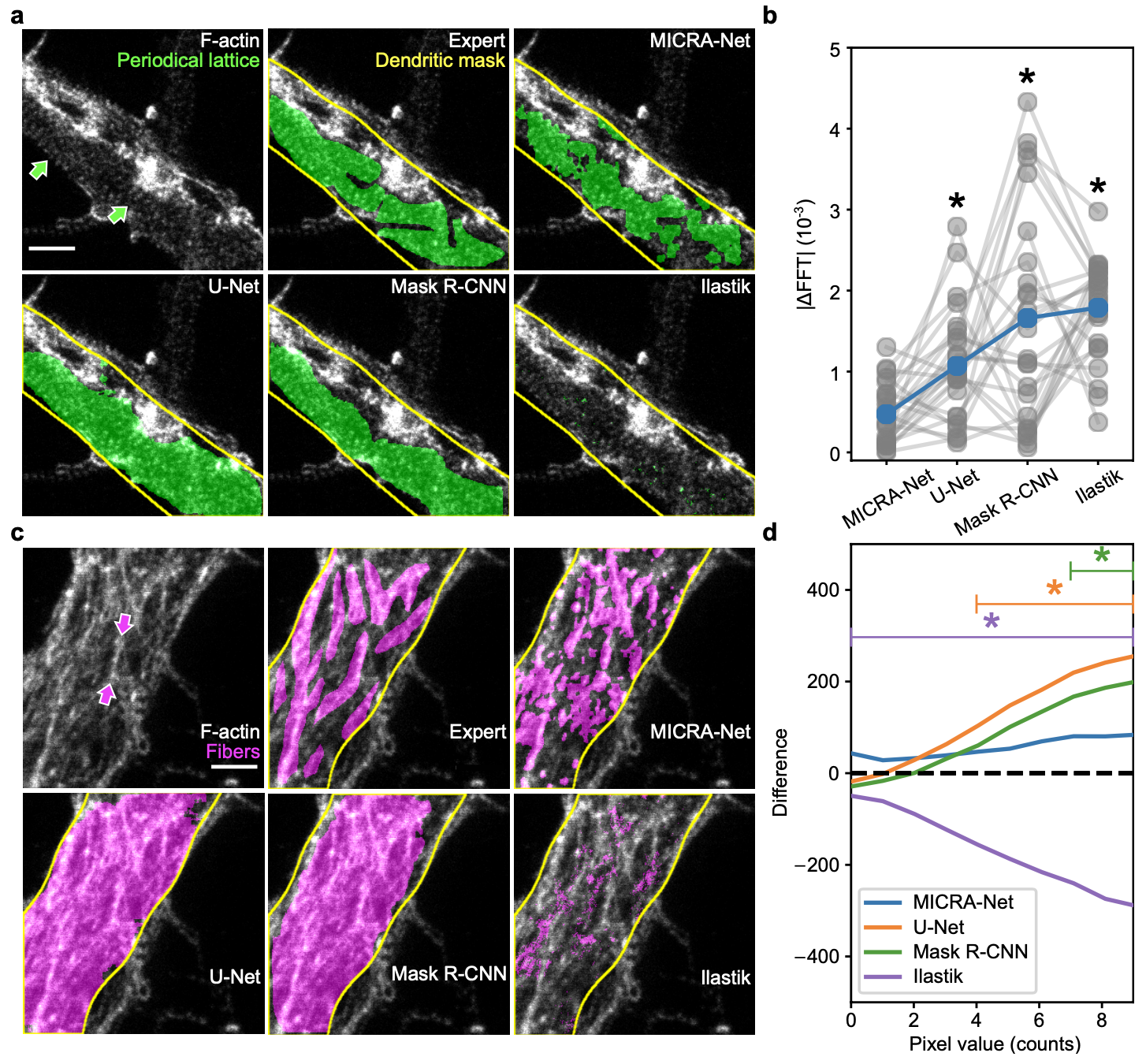
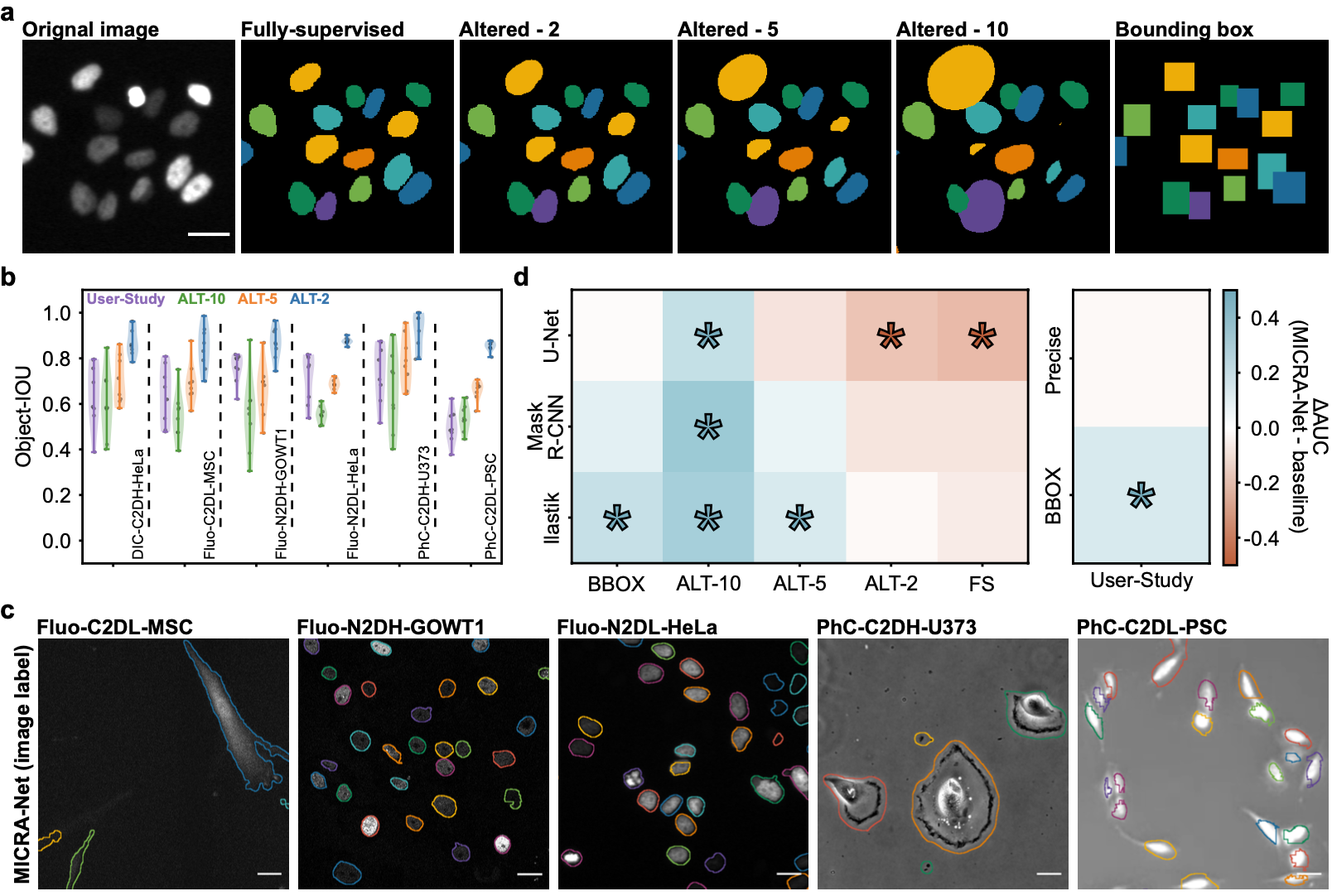
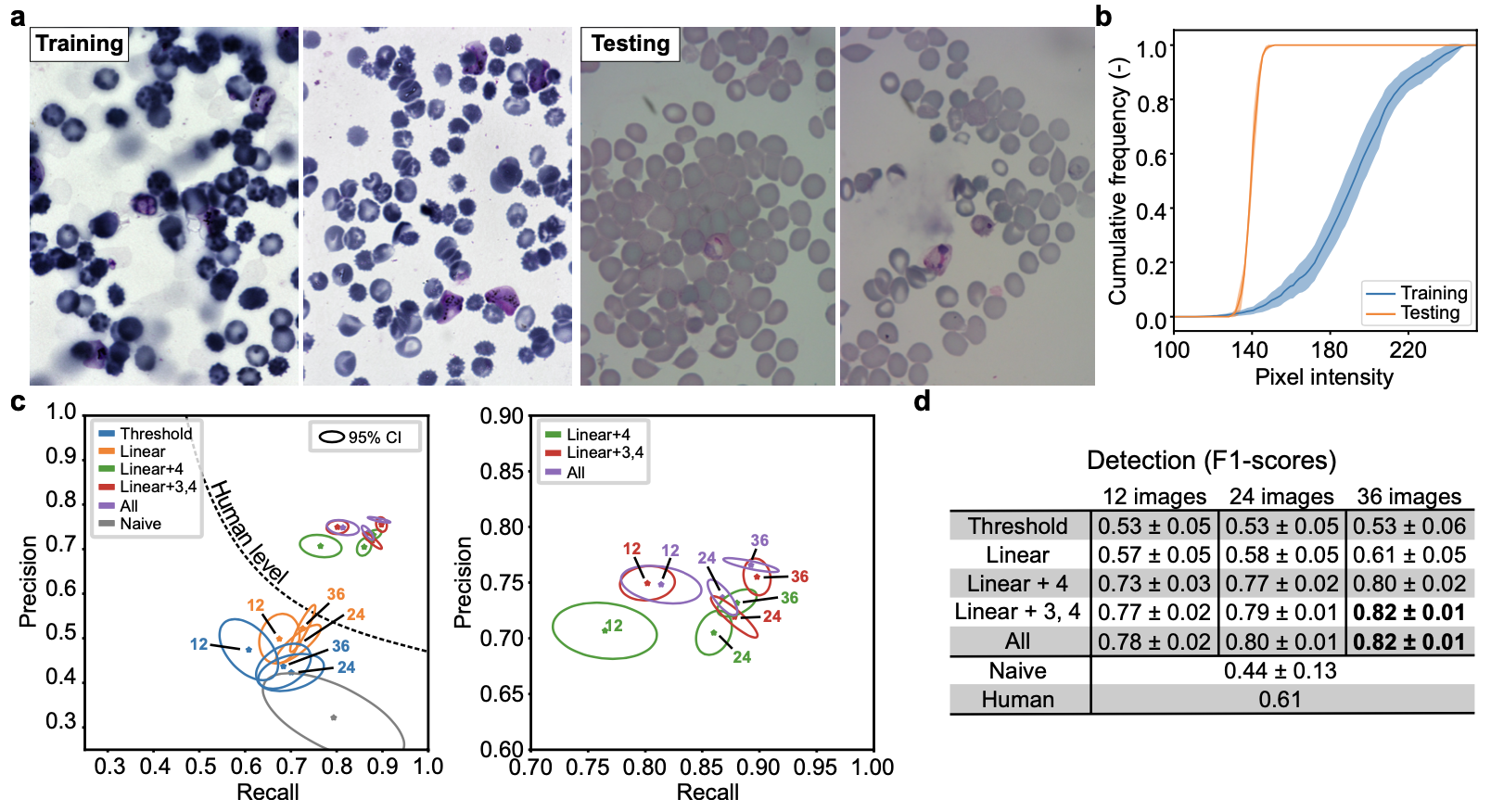
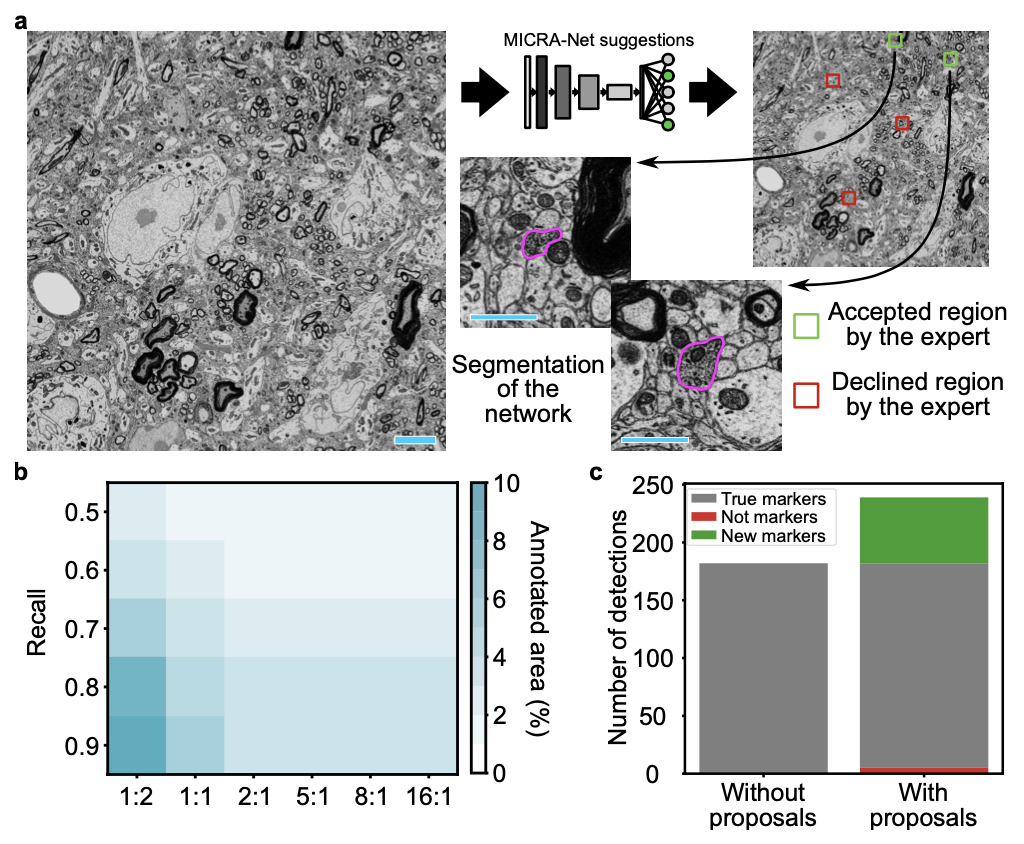
We provide the Ilastik zoo models in separate files to reduce the size of the download for the user. The Ilastik models are available from the following links : F-actin and Cell Tracking Challenge
Datasets
We provide the links to the datasets used in the paper below.
The F-actin dataset used for this paper is an in-house dataset. We release a public version of the dataset. If you use the provided dataset please cite the following paper: Neuronal activity remodels the F-actin based submembrane lattice in dendrites but not axons of hippocampal neurons. This dataset is available for general use by academic or non-profit, or government-sponsored researchers. This license does not grant the right to use this dataset or any derivation of it for commercial activities.
The Cell Tracking Challenge is a publicly available dataset. It may be downloaded from the Cell Tracking Challenge website.
The P.Vivax dataset is a publicly available dataset. It may be downloaded from the Broad Bioimage Benchmark Collection website.
The scanning electron microscopy dataset used for this paper is an in-house dataset. We release a public version of the dataset. If you use the provided dataset please cite us. This dataset is available for general use by academic or non-profit, or government-sponsored researchers. This license does not grant the right to use this dataset or any derivation of it for commercial activities.
Acknowledgments
Laurence Emond for F-Actin sample preparation and immunocytochemistry. Francine Nault, Charleen Salesse and Laurence Emond for the neuronal cell culture. Jonathan Marek and Renaud Bernatchez for the development of a custom Python annotation application. Thibault Dhellemmes for inter-expert axon DAB annotations in electron microscopy images. Christian Gagné and Marc-André Gardner for preliminary discussion on semantic segmentation. Annette Schwerdtfeger and Ana Gabela for careful proofreading of the manuscript. Funding was provided by grants from the Natural Sciences and Engineering Research Council of Canada (P.D.K. and F.L.C.), Canadian Institutes of Health Research (P.D.K.), CERVO Brain Research Center Foundation (F.L.C.), the Canadian Foundation for Innovation (P.D.K.). F.L.C. is a Canada Research Chair Tier II, Audrey Durand is a CIFAR AI Chair, and A.B. is supported by a PhD scholarship from the Fonds de Recherche Nature et Technologie.


